MCL1 Targeted Library
ChemDiv’s library of MCL1-targeting molecules comprises 12,000 compounds.
MCL1 (myeloid cell leukemia-1) is a widely recognized pro-survival member of the Bcl-2 (B-cell lymphoma protein 2) family, widely expressed in various tissues, and has been shown to be a promising target for cancer therapy. Primarily localized in the mitochondria, MCL-1 incorporates itself into the mitochondrial membrane through a hydrophobic tail. Its anti-apoptotic function is crucial for cell survival and homeostasis. Amplification and overexpression of MCL-1 have been observed in various human tumors, including both hematological malignancies and solid tumors.
Given the critical roles of BCL-2 family proteins in maintaining cellular homeostasis, perturbations in the interactions between pro- and antiapoptotic BCL-2 proteins, or in their expression levels, could disrupt the cellular equilibrium and lead to apoptosis resistance. Such imbalances contribute to the immortalization of cancer cells. To counteract these effects and control cell fate, novel small molecules have been developed to rectify the imbalance between pro- and antiapoptotic BCL-2 proteins, thereby restoring the apoptotic pathway. In recent years, there has been significant progress in the development of small-molecule MCL-1 inhibitors, with three MCL-1 selective inhibitors currently undergoing clinical trials [1].
The development of MCL1 inhibitors represents a promising field in drug discovery, particularly for the treatment of various cancers where MCL1 plays a critical role in cell survival and tumor progression. By specifically targeting MCL1, these inhibitors are capable of restoring the apoptotic balance within cancer cells, thereby facilitating programmed cell death and inhibiting tumor growth. The potential effectiveness of MCL1 inhibitors is underscored by their ability to address a key survival mechanism in cancer cells, offering a targeted therapeutic approach. This strategy not only holds promise for directly combating various cancers but also for overcoming resistance to existing therapies, making MCL1 inhibitors a focal point in the evolving landscape of oncological drug development.
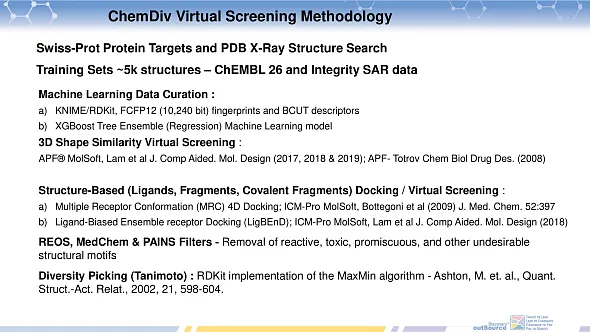
The development of the library using virtual screening methodology incorporated a sophisticated blend of machine learning data curation and various cheminformatics techniques, as detailed in the referenced slide deck. The initial step involved the use of KNIME, an open-source data analytics platform, in conjunction with RDkit, a cheminformatics software, to preprocess and curate the chemical data. This process included the standardization of chemical structures, removal of duplicates, and filtering of compounds based on specific chemical properties and criteria relevant to the target profiles. The library utilized FCFP 12, which are circular fingerprints capturing the molecular environment, and BCUT descriptors, which represent molecular properties related to atomic contributions to connectivity. These descriptors are crucial in capturing the essential chemical information of the compounds, aiding in the identification of potentially active molecules. The curated data and calculated descriptors were then analyzed using the XGBoost machine learning algorithm, a powerful and efficient implementation of gradient boosted decision trees. This model was trained on known active and inactive compounds to identify patterns and features that correlate with biological activity against the targeted molecular mechanisms. The development process also integrated other advanced methods, as described in the slide deck, which likely included additional statistical and machine learning techniques, as well as cheminformatics tools, to refine and optimize the screening process.
The library's development leveraged a comprehensive virtual screening methodology, integrating machine learning, data curation, and cheminformatics to efficiently identify and prioritize small molecules with potential therapeutic relevance. This advanced approach underscores the library's robustness and suitability for facilitating drug discovery initiatives.
References
[1] W. Xiang, C. Y. Yang, and L. Bai, “MCL-1 inhibition in cancer treatment,” Onco. Targets. Ther., vol. 11, pp. 7301–7314, 2018, doi: 10.2147/OTT.S146228.