Bromodomain Modulators Library
ChemDiv’s library of small molecule bromodomain inhibitors contains 5,816 compounds.
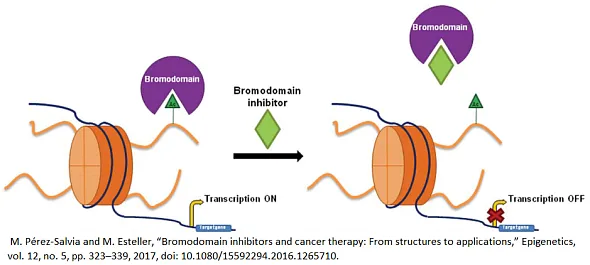
Bromodomains (BRDs) are evolutionarily conserved protein interaction modules that specifically recognize ε-N-lysine (KAc) acetylation motifs, playing a crucial role in the interpretation of epigenetic marks. These domains are integral components of a variety of larger protein complexes, many of which are key regulators of gene transcription. This includes histone acetyltransferases (HATs), ATP-dependent chromatin remodeling complexes, methyltransferases, and transcriptional coactivators, among others.
The fact that BRD-containing proteins are linked to many different diseases and that some histone deacetylase (HDAC) inhibitors have been shown to work strongly suggests that BRD inhibitors could be very useful in both drug discovery and academic research. The typically weak interaction between BRDs and their acetylated lysine substrates, the diversity and unique physicochemical characteristics of the acetyl-lysine binding site, and the extensive availability of crystal structures provide a solid foundation for the rational design of these inhibitors. This foundation aids in tailoring inhibitors that are specific, efficient, and capable of modulating the function of BRD-containing proteins, thereby opening new avenues for therapeutic intervention and mechanistic studies.
Small molecule compounds that inhibit BRD proteins play a pivotal role in drug discovery by offering a targeted approach to modulating epigenetic mechanisms that regulate gene expression. By specifically binding to BRDs, these inhibitors can disrupt the interaction between bromodomains and acetylated lysines on histone proteins, a key process in the reading of epigenetic marks. This disruption has the potential to alter gene expression patterns that are critical in the development and progression of various diseases, including cancer, inflammatory diseases, and neurological disorders. Because BRD inhibitors can precisely target these protein-protein interactions, they are useful for creating new therapeutic strategies. This opens the door to treatments that can selectively change the expression of pathological genes with possibly fewer side effects and better outcomes for patients.
ChemDiv introduces a novel library of BRD inhibitors or modulators, showcasing a curated collection of drug-like compounds specifically designed to modulate the protein-protein interactions of bromodomain-containing proteins with various pathways involved in critical physiological processes. This library has been thoroughly assembled, leveraging in-house structural biology insights, molecular dynamics simulations, virtual screening of ChemDiv’s proprietary chemical libraries, and medicinal chemistry filtering/ranking to refine the selection of potential hits. A thorough evaluation of the extensive structural data available on bromodomains has facilitated a comprehensive family-wide structural analysis of the human BRD family, assessing its "druggability." ChemDiv research teams have found promising chemical starting points for the creation of a targeted BRD inhibitor library by combining several in silico screening methods and using spatial information on possible acetyl-lysine mimetics. This strategic approach not only enriches the possibilities for drug discovery but also enhances the precision of targeting specific interactions within the BRD family. Our small molecule library of BRD inhibitors offers a significant advantage in drug discovery by providing a diverse array of compounds specifically designed to target and modulate the function of bromodomains, which are key players in the regulation of gene expression through epigenetic mechanisms. This targeted approach enables drug discovery researchers to efficiently identify and optimize potent inhibitors that can disrupt aberrant protein-protein interactions implicated in various diseases, including cancer and inflammatory disorders. Furthermore, the library's foundation on comprehensive structural and biological insights ensures a high degree of specificity and efficacy for potential therapeutic candidates, accelerating the pathway from initial screening to clinical development.