RNA Inhibitors: Small Molecules Targeting RNA | ChemDiv Libraries
Small Molecule INHIBITORS RNA: Therapeutic Targets, Mechanisms, and Validated Screening Libraries
Abstract: The pharmaceutical landscape is undergoing a structural shift from exclusive protein-targeting to the modulation of nucleic acids. Patent data and open innovation repositories (e.g., opnMe) confirm a significant increase in intellectual property regarding RNA modulation. INHIBITORS RNA—defined as small molecules capable of binding tertiary RNA structures or disrupting RNA-Protein Interactions (RPIs)—represent a rapidly expanding therapeutic class. This article details the scientific rationale, specific molecular targets (LIN28, TLRs), and the composition of specialized screening libraries available at ChemDiv.
1. The Scientific Paradigm: Drugging the "Undruggable" Transcriptome
Historically, RNA was considered an "undruggable" target due to the perception that it lacked deep, hydrophobic pockets characteristic of protein binding sites. However, recent structural biology data refutes this. RNA folds into complex secondary and tertiary architectures—including hairpins, internal loops, bulges, pseudoknots, and G-quadruplexes—that create distinct chemical environments suitable for small molecule binding.
Patent Landscape Validation
Analysis of the opnMe opnMINER document data confirms a surge in patent filings related to RNA modulation. This trend is driven by the necessity to target disease drivers that have no enzymatic activity or are "undruggable" at the protein level.
- Data Source: opnMe Document Data (ID: 41628982).
- Implication: The industry is moving toward direct modulation of mRNA translation and pre-mRNA splicing via small molecules (Inhibitors RNA), validating the commercial and scientific viability of this approach.
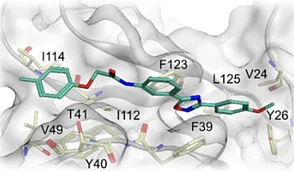
2. Mechanism Class I: RNA-Protein Interaction (RPI) Inhibitors
The functionality of RNA is largely dictated by its association with RNA-Binding Proteins (RBPs). Disrupting these complexes is a primary strategy for therapeutic intervention.
ChemDiv Library Specification: RNA-Protein Interaction Inhibitors
ChemDiv offers a focused library of >2,100 compounds specifically designed to target the protein-RNA interface. Unlike generic screening sets, these molecules possess vectors capable of interrupting the large surface area interactions typical of RPIs.
Key Validated Targets in this Library:
- LIN28 Inhibitors:
- Mechanism: LIN28 is a conserved RBP that binds to the terminal loop of the let-7 miRNA precursor, blocking its processing by Dicer.
- Pathology: Overexpression of LIN28 suppresses let-7 (a tumor suppressor), driving oncogenesis and maintaining stem cell pluripotency in aggressive cancers.
- Therapeutic Goal: Small molecules in the ChemDiv library are designed to bind the cold-shock domain or zinc knuckle domain of LIN28, restoring let-7 biogenesis.
- Toll-Like Receptor (TLR) Modulators:
- Mechanism: Several TLRs (specifically TLR3, TLR7, and TLR8) function as sensors for viral RNA. TLR3 recognizes double-stranded RNA (dsRNA), while TLR7/8 recognize single-stranded RNA (ssRNA).
- Therapeutic Goal: The library contains antagonists that block the binding of pathogenic RNA to these receptors, modulating the innate immune response and preventing autoimmune exacerbations.
- CRISPR-Associated (Cas) Protein Modulators:
- Mechanism: Cas proteins rely on guide RNA (crRNA) to locate DNA targets.
- Application: Small molecule inhibitors can provide temporal control over gene editing activity, reducing off-target effects by "switching off" the Cas-RNA complex.
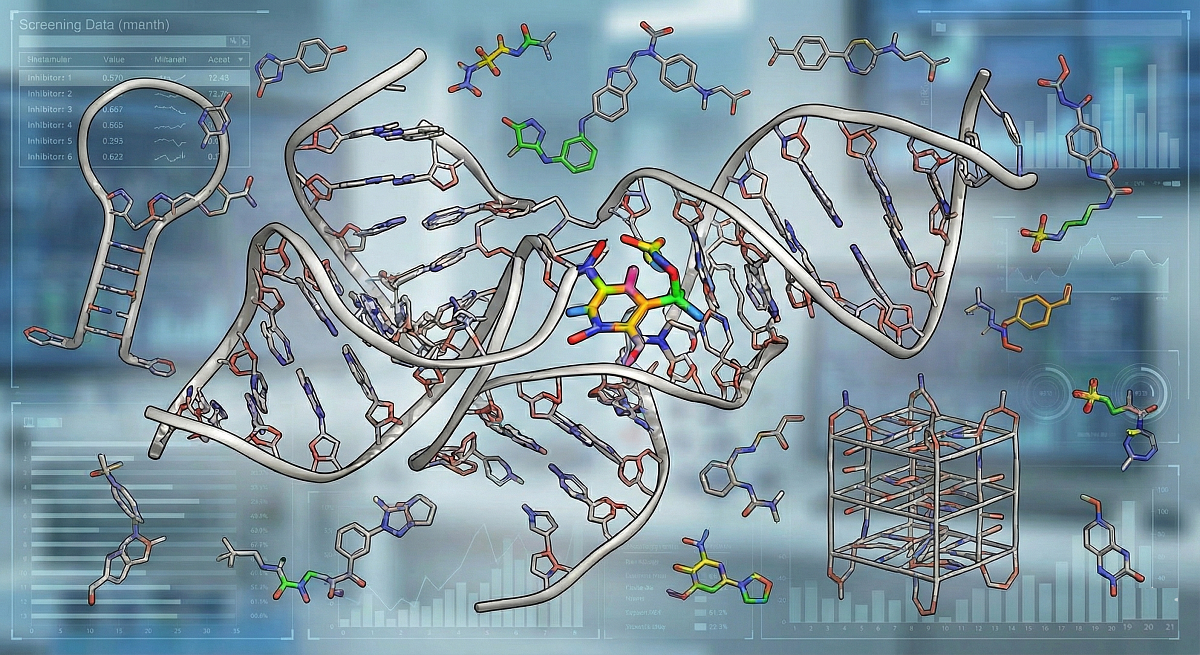
3. Mechanism Class II: Direct RNA Binding & Chemical Space
Targeting RNA directly requires a departure from "Lipinski's Rule of 5" compliant chemical space. RNA binders often require higher polarity and specific geometric configurations to engage with the negatively charged phosphate backbone and the hydrogen-bonding edges of bases.
ChemDiv Library Specification: RNA-Binding Chemical Space
- Volume: ~24,000 compounds.
- Design Methodology: This library is curated using Machine Learning (ML) algorithms trained on experimental data sets of known RNA ligands.
- Physicochemical Profile: The compounds are enriched for scaffolds that can stack with nucleobases (intercalators) or bind in the major/minor grooves of RNA helices.
- Structural Targets: The library is optimized to screen against:
- G-Quadruplexes: Four-stranded structures found in oncogene promoters (e.g., MYC, KRAS).
- Riboswitches: Regulatory segments of mRNA that bind small molecules to control gene expression.
4. Mechanism Class III: Targeting Repeat Expansions
Microsatellite repeat expansions are the genetic cause of over 40 neuromuscular and neurodegenerative disorders (e.g., Myotonic Dystrophy Type 1, Huntington’s Disease, ALS). These expansions result in toxic RNA species containing long repeats (e.g., CUG, CAG, GGGGCC) that sequester vital RBPs.
ChemDiv Library Specification: RNA Isosteric Trinucleotide Mimetics
- Volume: ~29,000 compounds.
- Scientific Basis: This is a first-in-class library of 3D-mimetics of trinucleotides.
- Mechanism of Action:
- Displacement: The small molecules mimic the structure of the RNA bases, binding to the toxic repeat RNA.
- Release: By occupying the repeat site, the compound displaces the sequestered RNA-binding proteins (such as Muscleblind-like proteins in DM1), restoring normal cellular splicing function.
- Selectivity: The mimetics are designed to distinguish between the pathogenic expanded repeats and normal cellular RNA.
5. Procurement and Screening Data
The availability of these libraries allows for immediate integration into high-throughput screening (HTS) campaigns.
| Library Name | Compound Count | Primary Mechanism | Key Targets |
|---|---|---|---|
| RNA-Protein Interaction | > 2,100 | Interface Disruption | LIN28, TLRs, Cas |
| RNA-Binding Chemical Space | ~24,000 | Direct Binding | Riboswitches, G-Quadruplexes |
| Trinucleotide Mimetics | ~29,000 | Structural Mimicry | CUG/CAG Repeats |
Actionable Step: Researchers and pharmaceutical developers can access structural files (SDF) and request formatting options directly through the ChemDiv portal.
Browse RNA Libraries at ChemDiv