ChemDiv has launched Kinome Profile—an advanced AI‑based binary classifier covering 50 key kinase targets.
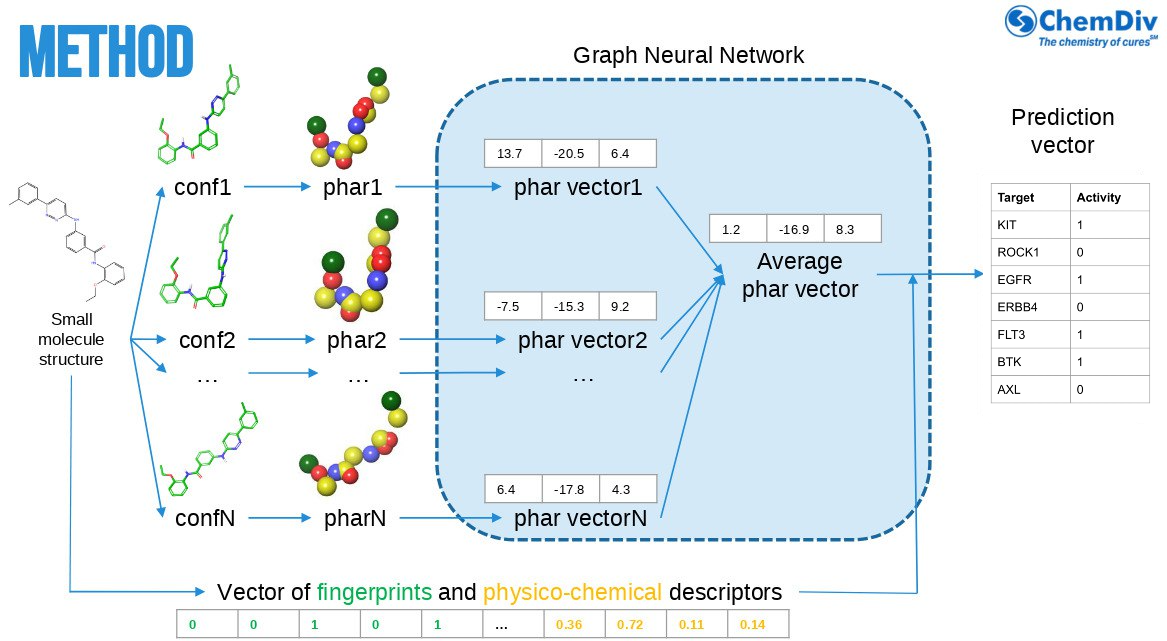
Kinome Profile is a 3D graph neural network that leverages 3D pharmacophore features, physicochemical descriptors, and Morgan fingerprints. Pharmacophores are derived from an ensemble of 3D conformers generated with a proprietary in‑house method.
Kinome Profile enables reliable, efficient predictions of kinase activity, helping teams save time and resources in drug discovery.
Coverage (50 kinases):
AXL, BLK, BMX, BRSK1, BTK, CAMK1D, CLK2, DAPK1, DDR2, EGFR, EPHA2, EPHA3, EPHB2, ERBB4, FES, FGFR1, FGFR3, FGR, FLT3, FLT4, FYN, GRK7, GSK3A, GSK3B, HCK, IGF1R, INSR, IRAK4, KIT, LCK, MARK1, MELK, MET, NEK2, NEK7, PAK2, PHKG2, PIM1, PIM2, PIM3, PLK1, PRKX, RET, ROCK1, ROCK2, SGK2, SGK3, TBK1, TYRO3, ZAP70.
Upload up to 250 structures in a single file to generate predictions across all 50 targets.
Further details are provided in the article.
We use cookies only to remember your preferences and provide better browsing experience. We do not sell user information. Here is our privacy policy.